| T H E N I H C A T A L Y S T | M A R C H – A P R I L 2001 |
|
|
|
Hot
Methods: Protein Profiling
|
by
Lance Liotta,
MD, PHD, NCI |
 |
|
NCI-CBER/FDA
Tissue Proteomics Initiative
|
Although DNA is the information archive of the cell, proteins do all the work. They operate through a complex network of interactions and post-translational modifications that cannot accurately be predicted by gene transcript profiling.
The proteome is not a static entity. It is different in each cell type and changes from one minute to the next, depending on the cellular microenvironment and the physiologic state of the cell. Thus, the proteomic challenge is much more than just cataloging all the proteins encoded by the expressed genes.
The true goal of proteomics—even grander than sequencing the genome—is nothing less than producing a complete wiring diagram of the protein networks of a cell, in health and under the influence of disease (1).
Tissue Proteomics Project
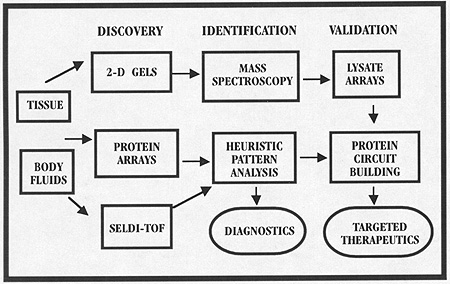
Three years ago, we established a joint research initiative between the NCI and the FDA called the Tissue Proteomics Project. The goal of this intitiative has been to originate and complete technology for studying proteomic networks and signal pathways in small quantities of microdissected human tissue cells directly from biopsy specimens.
In contrast to the expanding list of biotech companies and consortia moving into proteomics, our goal has been immediate patient-based clinical applications. We have originated a series of technologies to extract and analyze the pattern of proteins and determine the activation state of known signal pathways using microdissected human tissue or small samples of patient serum.
Our analytical tools are divided into two classes: protein microarrays that are used to profile the pattern of known proteins, and high-throughput biochips that can rapidly read out protein patterns, even if the identity of the proteins is unknown. We are using these tools to apply proteomics to samples from clinical trials and epidemiologic screening.
We believe advances in proteomics applied to tissue and body fluids will herald a new era in clinical research. Clinical investigators will monitor whole proteomic patterns of information, not just the concentration of one marker. Analysis of the patterns obtained before and after pharmaceutical treatment, or over the course of disease progression, could lead to insights about how an experimental drug works (or doesn’t work) for an individual patient’s disease and may reveal protein patterns that correlate with early disease or occult toxicity.
This article is part one of a two-part description of the proteomic tools under development in the NCI-FDA project. Here we will discuss "SELDI," a so-called protein biochip that can be used to rapidly generate protein molecular weight patterns—even though the protein identities are unknown. (Part two will discuss the use of protein microarray chips to monitor how drugs work in vivo.)
 |
|
Analaysis
of biological mixture by ablation, ionization, and time of flight
|
Part One: SELDI
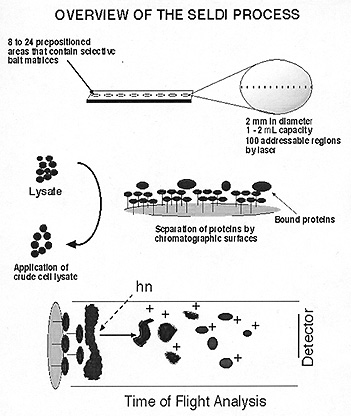
Surface-enhanced laser desorption–ionization time-of-flight mass spectrometry (SELDI-TOF) is a promising technique for rapid protein-pattern analysis of serum and microdissected tissue specimens. A large amount of protein-pattern information is generated from a small sample in a short period of time, and no labeling (for example, radioisotope or fluorescent) of the proteins is required.
SELDI starts with stainless steel or aluminum-based supports, or "chips," 1–2 mm in diameter. These are precoated with hydrophobic (reverse-phase), normal phase, metal affinity, and cationic or anionic baits. Solubilized tissue or body fluids in volumes as small as 0.5–l mL are directly applied to these surfaces.
After a wash step, proteins and peptides of selected affinity are retained on the chip and analyzed by mass spectrometry–time-of-flight technology similar to MALDI (matrix-assisted laser desorption and ionization). Ionized proteins and peptides are recorded as they strike the detector plate and, depending on their size, travel down the vacuum tube in a time-dependent fashion (larger molecules take longer).
Proteins are recorded as mass signature "peaks" and displayed as a standard chromatograph. Routine identification of the protein peaks seenwith this approach is not yet possible; that may change in the future, however, as the SELDI is coupled to instruments better adapted to protein microsequencing. Using SELDI, protein profiles can easily be obtained in minutes from as few as 25 to 250 cells. Furthermore, SELDI provides a complementary approach to 2D chromatography because SELDI is able to profile proteins regardless of their intrinsic hydrophobicity and has its best sensitivity (in the attomole range) to proteins below 15 kDa—a problematic size range for 2D-PAGE resolution.
Coupled to laser-capture microdissection (LCM)(2), SELDI protein profiling is an important tool for the molecular fingerprinting of cancer cells from human tissue, shedding light especially on changes in protein expression in early premalignant lesions.
Protein expression profiling is rendered as a traditional mass chromatograph or as a density graph "bar code" (see figure). We obtained reproducible patterns of protein expression from microdissected patient-matched cells that consistently changed over the course of the malignant process in esophageal, prostate, colon, ovary, and breast epithelium procured by LCM.
Intriguingly, these patterns showed evidence of cancer-type specificity.
Materials and Methods
![]() Tissue Preparation and Staining.
Frozen section slides, 8 mm thick, are prepared
as previously described (3),
except the proteinase inhibitor kit AEBSF (Boehringer Mannheim, Indianapolis)
is added to the staining baths at a final concentration of 2 mM to inhibit proteases.
With careful review of the histologic sections by a pathologist, each microdissection
should have a greater than 95 percent purity.
Tissue Preparation and Staining.
Frozen section slides, 8 mm thick, are prepared
as previously described (3),
except the proteinase inhibitor kit AEBSF (Boehringer Mannheim, Indianapolis)
is added to the staining baths at a final concentration of 2 mM to inhibit proteases.
With careful review of the histologic sections by a pathologist, each microdissection
should have a greater than 95 percent purity.
![]() Laser Capture Microdissection. Stained
tissue sections are subjected to LCM (Pixcell 100, Arcturus Engineering, Mountain
View, Calif.; see The NIH Catalyst, November–December 1997, "Hot
Methods"). Within five minutes of capture, microdissected cells are
lysed directly with 10 mL of an extraction buffer
containing 1% weight-to-volume (w/v) Triton-X-100 (Sigma, St. Louis), 1% (w/v)
MEGA 10 (ICN, Aurora, Ohio), 1% (w/v) octyl-b-glucopyranoside
(ESA, Chelmsford, Mass.), and 0.1% SDS (BIO-RAD, Hercules, Calif.) in a standard
1X phosphate-buffered saline (PBS).
Laser Capture Microdissection. Stained
tissue sections are subjected to LCM (Pixcell 100, Arcturus Engineering, Mountain
View, Calif.; see The NIH Catalyst, November–December 1997, "Hot
Methods"). Within five minutes of capture, microdissected cells are
lysed directly with 10 mL of an extraction buffer
containing 1% weight-to-volume (w/v) Triton-X-100 (Sigma, St. Louis), 1% (w/v)
MEGA 10 (ICN, Aurora, Ohio), 1% (w/v) octyl-b-glucopyranoside
(ESA, Chelmsford, Mass.), and 0.1% SDS (BIO-RAD, Hercules, Calif.) in a standard
1X phosphate-buffered saline (PBS).
SELDI Analysis. SELDI analysis is performed using an aliphatic reverse phase chip (H4 ProteinChipTM, Ciphergen, Palo Alto, Calif.). The bait surfaces on the chip are pretreated with 1 mL of acetonitrile (Sigma). Shortly before the acetonitrile completely evaporates, 1 mL of the lysate is applied to the bait surface. The analyte is allowed to concentrate by air-drying followed by washing two times for 5 minutes in 1 x PBS. Next, 0.3 mL of a saturated solution of 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid, Sigma, St. Louis, MO), the energy-absorbing molecule of choice, is applied to the washed surface of the chip and allowed to crystallize.
Reproducibility and Sensitivity of Protein Biomarker Profiling from Defined Cells. To assess the reproducibility of the molecular weight signatures generated by the SELDI protein fingerprint of LCM-derived cells, 1,500 cells of esophageal normal epithelium were microdissected, lysed, and applied to a predefined bait surface on a hydrophobic interaction C18 biochip (Ciphergen), as outlined above. A data set was generated using the cumulative detection of proteins from 12 of the possible 100 different addressable regions within a single biochip surface region.
Each of these experiments was performed in triplicate (three separate cumulative groupings of 12 different areas of the same spot), giving a total of 36 data points for each protein peak analyzed.
Different peaks and shoulders were chosen for their diversity in relative intensity to one another. These were analyzed using normalization to a protein that appeared consistently in all experiments.
The reproducibility of the detection of the tissue proteins was quantitatively analyzed by comparing the relative proportionality of a subset of these peaks with one another.
Analysis of the reproducibility of the protein profile obtained from several independent applications of equivalent loadings of the same lysate was performed using a lysate of microdissected esophageal normal epithelium from the same pertinent sample.
In this study, the lysate from 30,000 cells from the same patient was applied to 20 different C18 biochips with hydrophobic interaction bait surfaces (for an average protein load of 1,500 cell equivalents per bait surface) and analyzed in triplicate (12 regions per data set), for more than 720 data points per protein analyzed.
The coefficients of variance (less than 10 percent) and standard deviations generated from these analyses reveal that the molecular mass fingerprint of a given lysate can be reproducibly attained from any independent application.
We tested the sensitivity of protein detection by titrating lysates of microdissected normal esophageal epithelial cells from 1,500 to 0 cells/application. Each analysis was performed in triplicate.
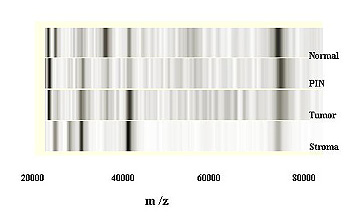 |
The results of this type of experiment can be represented as a "virtual gel"—a gel-like display that takes the data from the mass chromatogram and presents them as if they were from a standard 1D-SDS-PAGE gel "stained" for proteins, with the molecular-weight ranges displayed at the same scale as that seen in the chromatogram (see figure).
The lower limits of a reproducible protein fingerprint are in the range of 25 to 250 cells. When sensitivity is calculated as a product of cell-equivalencies, the detection limits become even more dramatic. This calculation is based on the fact that each individual bait surface on the protein biochip contains approximately 8,000 theoretically addressable regions, based on the area of a circle. Each of the individual protein profiles generated represents the cumulative detection of 12 different regions within one sample spot, so that the sensitivity of detection in terms of cellular equivalents is 12/8000th, or 0.15 percent, of the total lysate of the cells applied to the surface with-in one spot. This means that the biomarker protein profile in each reading represents the lysate of only two cell-equivalents!
Having achieved high sensitivity and reproducibility, SELDI was applied to microdissected prostate tissue—normal, premalignant, and invasive cancer from the same patient. It should be noted that these are microscopic lesions never before analyzed for their proteins. As shown in the figure, specific protein differences were uncovered, which were characteristic for each specific stage of cancer development, setting the stage for a new concept of molecular fingerprinting.
Pharmacoproteomics of the Future
In conclusion, protein biochips can be used to generate protein fingerprints from microscopic cell populations directly from human tissue. This technology has attained a high degree of reproducibility, sensitivity, and specificity.
Tissue protein profiling
may well become an essential component of pharmacoproteomics (patient-tailored
therapies), improving therapeutic assessment, drug or surgical intervention
strategies, toxicity monitoring, and disease diagnosis. ![]()
| "The Next Step: Exploring the Proteome," May 21, daylong, Masur Auditorium. Speakers from Large Scale Biology Corporation, Celera, University of Michigan Center for Proteome Studies, University of Geneva, Institute for Systems Biology, Carnegie Mellon University, Matrix Science, Proteome, Inc., and NCBI. Sponsor: Mass Spectrometry IG and many institutes. Info: Sanford Markey 496-4022. |
References
1. L. Liotta and E. Petricion. "Molecular profiling of cancer." Nat. Genet. Rev. 1: 48–56 (2000).
2. C.P. Paweletz, J.W. Gillespie, D.K. Ornstein, et al. "Rapid protein display profiling of cancer progression directly from human tissue using a protein biochip. Drug Devel. Res. 49: 34–42 (2000).
3. N.L. Simone, A.T. Remaley, L. Charboneau, et al. "Sensitive immunoassay of tissue cell proteins procured by laser capture microdissection." Am. J. Pathol. 156: 445–452 (2000).
| Instant Proteomics? |
|
Just when you were getting the hang of potato chips, computer chips, and cDNA chips, along comes proteomics, and, naturally, the ProteinChipTM—arrays of chromatographic spots to help you analyze proteins, in much the same way oligonucleotide microarrays allow you to look at gene expression. So far, about six NIH labs have forked over the $145,000 to acquire the ProteinChipTM reader, says Ciphergen rep Gerard Hoehn, who spoke at NIDCR on March 8. Depending on how busy a lab keeps the reader, it may also burn its way through $30,000 to $70,000 worth of chips in a year. Hoehn estimates that, worldwide, about 60 scientific papers have now been published about work that used the Ciphergen chips. Hoehn said the chips could be used to accomplish a mouthwatering menu of protein-analysis tasks. The most crude would be separating a mishmash of 500 to 750 proteins from any complex biological sample on a 12 x 8 array. With samples of a mere 2,000 cells, researchers could thus go on a reasonably fast-paced fishing expedition for biomarkers—proteins whose characteristics (concentration, molecular weight, or binding properties) are altered in response to some event—a disease, toxin exposure, or age. Closely related to a biomarker hunt would be protein expression profiling (see "Beyond Genomics to Clinical Proteomics")–using the chips to compare differences in protein expression in different cell types. Analysis of signal transduction is a hot basic science area for protein chips, Hoehn said, including examination of critical posttranslational modifications of signaling molecules and identification of ligands for orphan receptors. They can also be used to detect and, with some extra fiddling, locate various "ations" on a protein—phosphorylation, glycosylation, ubiquitination—any post-translational modification that alters molecular weight. Before the chips can do their magic, they must first be "derivatized"—have their surfaces coated with an array of chemicals or covalently bound biological molecules, or baits, that will differentially trap proteins in a sample. Inorganic toppings come preloaded onto chip arrays (again, see main article). Biological toppings must be selected and attached by the investigator to a carbonyl diimidazole- or epoxy-coated chip. Possible additions could include antibodies, receptors, ligands, other proteins, or even DNA. NIMH’s Brian Martin, collaborating with FDA’s Li-Shan Hsieh, has been test-driving the ProteinChipTM Platform—a machine on intermittent loan from the company. He finds it reliable, exciting, fun, and a pain, all at once. "There’s no shortage of data. The problem is designing experiments so you can interpret the data and have [them] mean something," Martin says. He and Hsieh are looking for allergens in latex. Martin’s also going after some putative transcription factors and receptors for scorpion toxins. |