



Assembly of large protein complexes is always amazing, but the filament assembly is especially so because most of the process takes place outside the cell. Flagellin subunits, made inside the cell, flow through a channel in the hook and filament and assemble at the distal end of the elongating filament just at the cap.
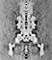
Using low-dose electron cryomicroscopy, we obtained images of filaments, hooks, and motors that we had isolated from cells. The micrographs are relatively noisy, but by averaging many images, we produce clean, three-dimensional maps that reveal the subunit and domain organization of the component proteins. We now believe the motor consists of three rings of subunits: the stator, which conducts protons; the rotor, or M-S-ring; and the C-ring, which could be part of either rotor or stator. Our maps suggest the hook and filament both possess a 30-Å-diameter protein export channel, suggesting that the proteins forming the filament must pass through the channel in an unfolded state.
A: About 15 years ago, I began work on the bacterial flagellum in collaboration with Lucy Shapiro at Stanford University in Palo Alto, Calif., who was working on the developmental aspects of flagellar biosynthesis. Shapiro believes that we must recognize structural organization as a key element in our understanding of the developmental program of an organism. I have focused on visualizing the isolated flagellar components in order to understand how they fit together and act coordinately to produce function.
We began with the hook and motor and then undertook studies of the filament. Recently, we shifted from simply obtaining three-dimensional images to assigning protein sequences to structural features. We have pushed the resolution of the filament to about 10 Å, which permits us to visualize alpha helices. For the motor, we have been trying to locate the components that are known, from other genetic studies, to generate torque. We also want to visualize the export apparatus that directs flagellin subunits into the flagellar channel.
Q: Which findings have been most surprising to you or to other scientists?
A: We were all amazed and delighted by the intricacy of the flagellar structures, especially the motor. The proteins are not compact blobs, but instead fold into exquisite shapes. The L-ring protein, for example, looks like an upside-down letter J. The filament also has an unusual concentric tubular structure built from alpha helices. The inner cylinder is the export channel. I wonder whether the character of the protein side chains that face into the lumen is essential to the flow of exported subunits.
Q: What were the greatest stumbling blocks, and what new observations, techniques, reagents, or insights helped you to get past them?
A: It was a challenge to find gentle methods for separating the flagellum from cell debris while retaining the torque-generating proteins. The availability of antibodies was essential to demonstrating that we had been successful in this. The relationships of the export proteins to the flagellar structure remain mysterious. A battery of antibodies would simplify detection of these proteins in our preparations. If the proteins are present, then immunomicroscopy would help localize them to flagellar features.
Q: Do you see any potential areas where this research might provide insight to clinical scientists?
A: Virulence in bacteria such as Shigella flexneri -- a cause of bacterial dysentery -- requires an export apparatus that appears homologous to that of the flagellum. Structural studies of the flagellum and its export apparatus might suggest ways to attack the mechanism by which bacteria export pathogenic proteins.
Q: How are you following up on this work, and what questions would you ultimately like to answer?
A: We are trying to extend our structural studies of the filament and hook to yet higher resolution, where we might be able to trace the peptide chain. We also want to obtain images of the complete motor and export apparatus and to assign each feature to a particular polypeptide sequence. We would especially like to see what structural changes cause the motor to reverse its direction of rotation causing the bacterium to change directions as it swims.
I am indebted to my collaborating colleagues at Brandeis, Noreen Francis, David Morgan, Gina Sosinsky, and Dennis Thomas; those at Teikyo University in Utsonomiya, Japan, Shin-Ichi Aizawa and Kenji Oosawa; and others at Yale University in New Haven, Conn., Robert Macnab and members of his laboratory. This work was supported by NIGMS.