| T H E N I H C A T A L Y S T | S E P T E M B E R – O C T O B E R 2006 |
|
|
|
New
NIAID Program with Universal Utility
|
by Jason Bardi |
 |
|
Signaling
Network
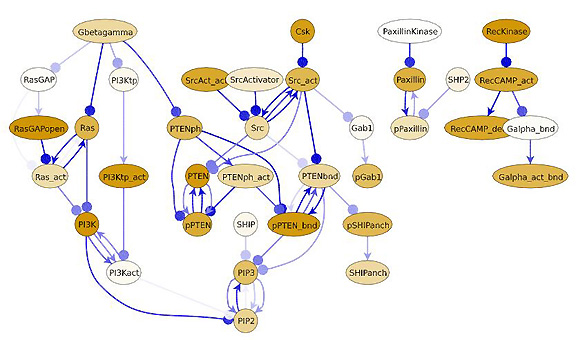
[for chemosensing] generated by mathematical modeling
software that carries out spatially resolved simulations of the behavior
of a given network when it is stimulated or perturbed. [See
additional figures below.]
|
About five years ago, NIAID scientist Ronald Germain wrote an article for Science magazine with an unlikely title—"The Art of the Probable."(1) He argued in favor of a new, broader way of looking at the immune system—more through the eyes of an engineer or a mathematician than those of a biologist.
Advocating such an approach was perhaps unusual for a classically trained immunologist like Germain, who is deputy chief of the NIAID Laboratory of Immunology and chief of the Lymphocyte Biology Section.
But perhaps it was his not being a physical scientist that allowed Germain to appreciate what mathematics might bring to his field—especially then, he recalls. In 2001, the draft of the complete human genome had just been published, and biology was awash with genomic and related data. The field of immunology was going through a golden era of discovery, and many of the molecular and cellular players of the immune system were known. But the connections between the parts that had been catalogued—and deep insights into how this machinery produced immune phenomena—remained elusive. Germain observes today, "We are still far from really understanding the operation of the immune system, by which I mean being able to predict and finely manipulate its behavior."
A broader understanding of immune physiology, he says, might come from embracing a more comprehensive and quantitative approach—an approach broadly encompassed by the otherwise overused term "systems biology." This is a way of studying biology that involves obtaining detailed information on enough components of a system (or even the entire system for simple organisms) to allow quantitative modeling of complex in vivo molecular and cellular events, such as pathogen-induced disease or immune system responses in mice or humans.
To that end, Germain will be leading a new intramural program that will combine systems level analysis with mathematical modeling to better understand host defenses and immune pathology—the Program in Systems Immunology and Infectious Disease Modeling (PSIIM).
Systems Biology Basics
Systems biology is a way of asking how large systems of molecules, cells, and tissues interact with each other. In the past few years, numerous studies have revealed the presence, actions, and interactions of many of the genes, proteins, lipids, carbohydrates, and other molecules in healthy and diseased tissues.
But knowing the molecular players may not be enough to translate these research results into health benefits. How the pieces fit together to generate what we observe as cellular or, on a higher scale, organismal behavior may depend on many variables, such as their concentration and location in the cell and the signals the cell receives from its environment, Germain notes.
Furthermore, even if all these variables are known, predicting what will happen to the system over time is complicated by the fact that the interactions are numerous and non-intuitive, and they often result in nonlinear behavior. The number of relevant components and interactions, he observes, surely exceed the oft-quoted limit of seven items that the average human can hold in short-term memory at one time.
The only tools equal to the task are mathematical and machine based—allowing us to translate hypotheses about the systems of interacting molecules and cells into formal descriptions and then equations that can be evaluated quantitatively with the help of computers.
But the requisite level of expertise in applied mathematics and computer science is beyond the scope of most biology laboratories.
"Biologists often think about being more quantitative and trying to model their system of interest explicitly, but they would like to do this without having to become expert in the mathematics and computer scripting needed to do such modeling at a sophisticated level," Germain says. "Having access to a resource such as a strong, well-staffed systems-biology program can help make that happen."
 |
 |
|
PSIIM
Director
Ron Germain hoping to bring quantitative analysis and computer modeling
to mainstream immunology
|
Current
PSIIM Crew:
(left to right) Frederick Klauschen, postdoctoral fellow; Martin Meier-Schellersheim,
staff scientist and creator of Simmune; Alison Wise, postbac student;
and Bastian Angermann, physics Ph.D.
student in a joint program with the University of Hamburg, Germany, and
the PSIIM
|
Biologist Meets Theorist
When he published his Science article in 2001, Germain was well aware that his own expertise was not in mathematics and computer science. But when a colleague introduced him to Martin Meier-Schellersheim, a physicist who had spent his time as a Ph.D. student in Germany working on ways to create computational programs to model biological systems, Germain saw the perfect opportunity to move his ideas forward.
"It was very clear that Martin’s goals were absolutely congruent with what I wanted to do," recalls Germain. "And what he wanted was to be in a place that had the biology to interact with the software tools that he would create."
The two began working together when Meier-Schellersheim came to NIH as a postdoctoral fellow in the Laboratory of Immunology in 2001. Over the next several years, as Meier-Schellersheim continued improving his modeling and simulation software, Germain worked with the leadership of NIAID to develop plans for a formal program in systems immunology built around the advances in computational biology that were being developed in the lab. NIAID Director Anthony Fauci and then–acting NIAID Scientific Director Kathryn Zoon expressed substantial interest in such a program—which was reinforced in 2005, when NIH Director Elias Zerhouni and Deputy Director for Intramural Research Michael Gottesman sat down with Germain and Meier-Schellersheim to discuss systems biology and the work of the laboratory.
Zerhouni asked probing questions on topics such as whether the new software Meier-Schellersheim had developed dealt adequately with stochastic behavior. After getting what were clearly satisfying answers, Zerhouni expressed great interest in the research program being built around the new software approach, given its congruence with extramural Roadmap Initiatives to which he had made a strong commitment.
The end result of all these discussions and interactions was the recent decision by Zoon, now newly appointed director of NIAID’s Division of Intramural Research, to identify the resources needed to establish the PSIIM.
The immediate goal of the PSIIM will be to use a systems approach to better understand the complex biochemical networks that regulate the interactions between infectious organisms and the human or animal cells they infect.
The program will use state-of-the-art experimental approaches to determine how closely simulations of biological models can explain and predict real behavior and improve the models when they fail to match reality. As the models improve, scientists will eventually gain the ability to predict how drugs and other interventions will affect a cell or organism and whether the host will tolerate them.
Although most of the studies will be conducted with ordinary infectious pathogens, special facilities in the new C.W. Bill Young Center for Biodefense and Emerging Infectious Diseases will enable PSIIM scientists to examine such questions with microbes that cause diseases such as anthrax, virulent forms of influenza, tularemia, and plague.
The PSIIM approach will allow scientists to seek more complete answers to key questions that could not be addressed at this scale even a few years ago—such as how infectious organisms invade human cells, how the toxins they produce cause cell and tissue destruction, and how these pathogens evade or manipulate the immune response.
"Once we understand these interactions in a more systematic way, we can make strategic decisions about how to interfere with infectious disease pathology or how to direct immune responses to better fight infections," says Zoon, adding that these new insights can serve as the starting point for the design of new drugs to treat diseases or the development of new vaccines.
When the Immune System Goes Awry
Likewise, PSIIM will allow scientists to ask how perturbations in some of the same networks that defend against infectious agents lead to autoimmune diseases, which arise when the immune system goes awry.
One example Germain discussed in his 2001 Science article is multiple sclerosis (MS), a disease that usually first appears in adults between the ages of 20 and 40 and manifests in intermittent episodes of an array of impairments, such as loss of balance and muscle control, numbness and pain, and compromised vision and hearing.
MS is characterized by the presence of autoreactive T cells that recognize, attack, and destroy myelin sheaths within the central nervous system. The resulting disruption of the neuronal transmission of electrical impulses gives rise to the symptoms.
Much is known about MS etiology, "and we have a good idea of the nature of pathogenic cells that are present in people who have the disease," says Germain, but many unanswered questions remain. For example, how does the disease start? Autoreactive T cells undergo clonal expansion during an immune response, rapidly mutiplying until their numbers are thousands of times greater in diseased than in normal individuals. But how many cells does it take to start the process ultimately leading to tissue damage?
"The answer could be as few as one," given the explosive growth potential of T lymphocytes and the feed-forward effects that accompany the initial expansion and production of immune effector cells that damage tissue, Germain notes. "But what are the conditions that allow that first cell to respond and to initiate a process that eventually breaks through the many checkpoints that have evolved to prevent such autoimmunity?" By creating computer models able to simulate the biology of cells, tissues, and, eventually, organisms, Germain and his colleagues hope to answer questions like these.
Enter Simmune
One of the main tools for addressing these issues is a software package called Simmune, through which researchers may easily make, modify, and run simulations of detailed quantitative models of the biological systems they have studied in their labs for years.
This software package, created by Meier-Schellersheim and his associates, allows a scientist to use a simple graphical interface to define the interactions between pairs of molecules. These are the building blocks for signaling networks that when activated can produce hundreds or thousands of multimolecular complexes. The software automatically creates mathematical representations of these resulting networks from this simple molecular binding information. Once the scientist adds quantitative information on association/dissociation rates, enzymatic transformation rate, and the like that come from laboratory measurements, Simmune allows the researchers to investigate complex signaling activities within simulated cells without ever having to work with the underlying equations.
Before Simmune, making such mathematical models by hand could take months and required extensive expertise in applied mathematics. Even making changes to an existing model was very time-consuming, limiting the extent of what could be modeled.
"One of the great advantages of Simmune is that it gives biologists a way to do the difficult mathematics needed for such modeling without having to actually be involved with the mathematics," says Germain. "The hope is that these models will provide a deeper understanding of how complex behaviors arise, leading to new insights into disease."
Test Cases
In the past year, a beta version of the new software has been developed and tested; Germain and Meier-Schellersheim expect to release this version for academic purposes this fall.
In a well-air-conditioned basement computer laboratory on the NIH campus this summer, Meier-Schellersheim demonstrated the new software to a visitor. He clicked a few buttons and built a "molecule," which resembled a simple version of the old ball-and-stick models from introductory chemistry classes. One end of the molecule was designated the binding spot for another molecule that was also encoded using this simple interface.
"This particular end of the molecule can also be defined in the software as binding to additional molecules, providing an easy way to specify the complex set of interactions a single protein can engage in," said Meier-Schellersheim. He explained that once the numerous interacting components are entered, along with relevant data such as their concentrations and distribution, the software then automatically builds the equations that allow a simulation of biological acxtivity to take place.
Meier-Schellersheim, Germain, and their colleagues described the first stringent test of Simmune this summer in the journal PLoS Computational Biology.(2) Showing that the new software can accurately predict cell function in both time and space, they used Simmune to model a complicated cell-biological behavior known as chemosensing—a fundamental biological process whereby cells sense and respond to external signals, such as inflammatory chemicals involved in an immune response.
They modeled what happens in a stimulated cell to the distribution of a membrane-associated phospholipid. The concentration of the phospholipid changes during chemosensing mainly due to the action of two enzymes that synthesize or break down this molecule.
Contrary to previous thinking that one unknown cell-wide mechanism exerts different effects at different cell sites, the Simmune model predicted that the enhanced concentration of phospholipid at the "front" end of the cell (facing the source of chemical signals) resulted from a combination of two known mechanisms—a very rapid local inhibitory activity and the slower movement of another molecule to a distant part of the cell.
In collaboration with Tian Jin and Xuehua Xu of the Laboratory of Immunogenetics, NIAID, the team tested its Simmune-generated predictions in the laboratory and found a very close match with the experimental data.
The real power of the software, Meier-Schellersheim adds, is that it can be applied to nearly any cell-based biological system. "This is a tool that can simulate signaling and cellular processes in general," he says, "whatever system you are interested in."
And the real strength of the PSIIM program, Germain observes, comes from the connections it will help establish among scientists throughout and beyond NIH.
At the moment, though, the core PSIIM team is still being assembled. Germain is working on recruiting team leaders to head up groups in areas such as bioinformatics and proteomics, which will eventually be core parts of the PSIIM.
"We are very hopeful
that what we have started in our institute will build out as a trans-NIH initiative,
with strong links to the extramural community as well," says Germain. "The
ultimate goal is not only to solve important problems in immunology and infectious
disease within our group, but also to establish a large network of teams engaged
in creating the quantitative tools and computational methods for all biomedical
investigators to use in the coming years." ![]()
1. R. Germain, "The art of the probable: System control in the adaptive immune system," Science 293, 240 (2001).
2. M. Meier-Schellersheim, X. Xu, B. Angermann, E.J. Kunkel, T. Jin, and R.N. Germain, "Key role of local regulation in chemosensing revealed by a new molecular interaction-based modeling method," PLoS Comput. Biol. 2(7), e82 (2006).
 |
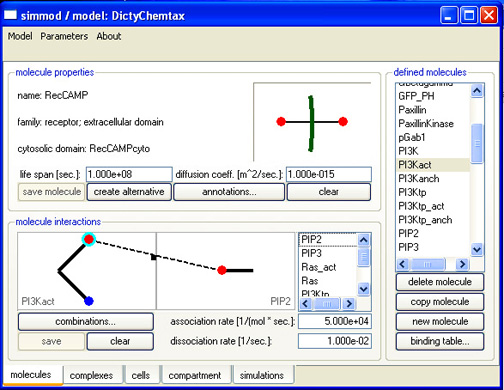 |
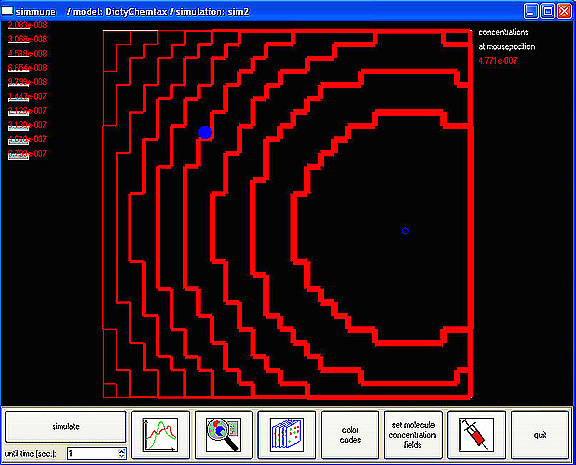
| Gradient2 shows a computer-generated gradient of chemoattractant (red lines), the origin of the gradient (blue open circle), and a single "synthetic" cell created in the modeling program (filled blue circle), whose response to the surrounding gradient is simulated by the program over time (see the other legends) | MolScreen shows a screen shot of the interface of Simmune used to enter/describe the interactions of molecules that have been created in Simmune. This simple click-and-drag process is the fundamental step in using Simmune to simulate signaling networks; it provides the information the software needs to automatically generate a model of the signaling network involving these interactions and to carry out spatially resolved simulations of the behavior of the network when it is stimulated or perturbed.ĘThe image Signaling Network above was generated by the use of the MolScreen entry process. |
 |
|
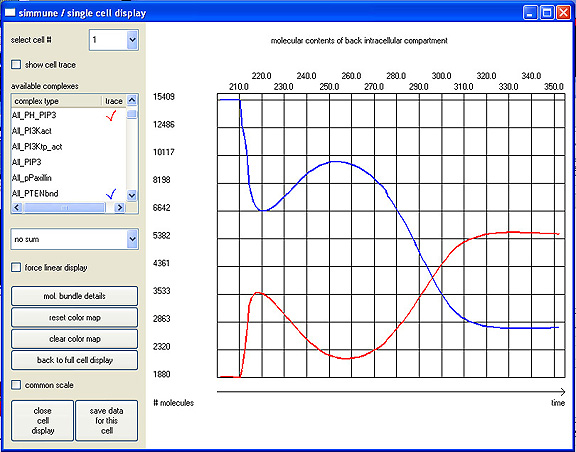
SingleCell1 shows the results of running a simulation of one synthetic cell with the Signaling Network pictured above, following exposure at time 210.0 seconds to a chemoattractant gradient. The blue curve shows the concentration change over time of the lipid PIP3 at the "back" of the cell after exposure to the gradient, that is, in the region of the cell experiencing the lowest concentration of attractant. The red line shows the change in the phosphatase PTEN in the same region of the cell over the same time period. The results reveal the oscillating nature of the changes early after chemoattractant contact, an unexpected result the PSIIM team confirmed by singe-cell imaging in collaboration with T. Jin and X. Xua of the LIG, NIAID. |