| T H E N I H C A T A L Y S T | S E P T E M B E R – O C T O B E R 2003 |
|
|
|
On
the Road to Crystal Clarity
|
 |
|
from published works by NCI's Sriram Subramaniam and
colleagues in Nature Structural Biology, August 2002, and the Journal
of Bacteriology, March 2003.
|
|
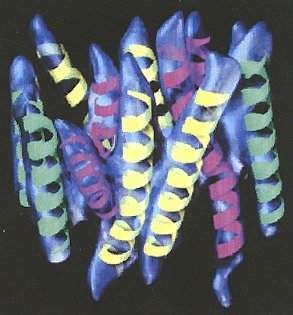
Three-dimensional
density map of the oxalate transporter, with the 12 idealized helices
grouped into three sets and superimposed manually on the map. Those colored
green are nearly perpendicular to the plane of the membrane, those colored
yellow contain bends in the transmembrane region, and those colored magenta
contain both a bend and curve in the transmembrane region.
|
|
Question: What’s complex, wiggly, mysterious, not very abundant, hard to pin down, risky, essential for life—and something NIH is getting to know better than anyone else in the world? A. slime molds B. giant amoebae C. policy on stem cell research D. membrane proteins Answer: D |
Although membrane proteins were once thought impossible to crystallize, their structures are now being published routinely and understood—at least a bit—and NIH’s intramural research program has something to do with this progress. According to NINDS’ Joseph Mindell, NIH now has "[one of] the strongest groups of membrane protein crystallographers [and] membrane protein structural biologists there is."
These investigators are exploring new techniques for the analysis of membrane proteins and are applying new developments to understand how membrane proteins work. Spread throughout NIH’s institutes and centers, these scientists are increasingly confident of their abilities. "Our view is, once you have enough of the membrane protein, you can do the structure," says NIDDK’s Susan Buchanan.
Membrane Protein Mysteries
Estimated to comprise about one-third of all proteins, membrane proteins act as critical gatekeepers to the cytoplasm. Some convey signals across the phospholipid bilayer in response to conditions outside the cell by changing their shape. Other membrane proteins transport vital small molecules, frequently in a regulated manner.
Essential to the life of the cell, membrane proteins are also increasingly essential to medicine. Between 40 and 60 percent of pharmaceuticals today target membrane proteins. Astonishingly, however, the fundamental features of most of these clinically important targets, including their structure and function, are poorly understood.
Low expression level, large size, multimeric composition, and complex arrangement of domains conspire to make membrane proteins notoriously troublesome to study. Unlike cytoplasmic proteins, membrane proteins contain very large hydrophobic regions, making purification difficult. The crystal structures of only a handful of mammalian membrane proteins have been solved.
In the past, scientists could expect to spend a decade divining the optimum conditions for the production, purification, and crystal formation for a single membrane protein.
But the times are changing. Over the past five years, scientists here and elsewhere have solved the structures of nearly 80 bacterial pumps, channels, and receptors. It now takes just one to three years to form crystals and analyze data for some bacterial proteins—just about right for a postdoc project, Buchanan points out.
A sizeable number of NIH scientists are now riding this wave of discovery. The NIH Catalyst interviewed seven scientists to survey the range and depth of structural membrane protein discovery at NIH.
From
Expression
To
Crystal Structure
And
Protein Function
Before scientists can solve the structure of a membrane protein, they have to be able to express it in quantities sufficient for their method of structural analysis. If that method is crystallography, the quantities required are large and the challenges of producing these quantities of usually scarce molecules are huge. Experts here say one of the keys to NIH’s lead in analysis of membrane proteins is its growing savvy in expression techniques.
Advances in the expression of membrane proteins have made possible not only crystallographic analysis of those proteins but also their study by complementary techniques.
 |
|
Susan
Buchanan
|
Iron Transporters
With more than 15 years of experience solving the crystal structures of membrane proteins, Susan Buchanan, an investigator in NIDDK’s Laboratory of Molecular Biology, observes that there are still a "huge number" of technical developments needed to optimize the expression and purification of membrane proteins. She and other NIH scientists are hammering away at these needed advances.
Buchanan focuses on iron transporters from several gram-negative bacterial species. Iron is necessary for bacterial proliferation; if iron uptake could be blocked, she observes, an infection could be stopped in its tracks.
"Pathogens such as Neisseria meningitidis and Yersinia pestis have completely different iron transporters from [Escherichia] coli, and that’s a good thing to study structurally," Buchanan notes, also pointing out that these transporters are highly antigenic and probably good vaccine targets.
Iron transporters have other important qualities. Energy is required for transport, and it appears that many different outer-membrane iron transporters are coupled to a common inner-membrane protein involved in energy transduction. Bacterial iron transporters are picky about their passengers, usually ferrying iron bound to carrier proteins rather than lone iron atoms. In fact, they are highly specific for proteins binding Fe(III).
Buchanan’s first crystal structure of an outer-membrane iron transport protein yielded not only a smaller-than-expected 22-stranded transmembrane b-barrel (all predictions at the time suggested 30 or more b-strands), but an additional surprise as well–a globular domain sitting inside the pore.
This globular domain is thought to play roles in ligand-binding specificity and interaction with the inner-membrane protein involved in energy transduction.
"So we view the barrel as a rigid scaffold that allows the rest of the protein to be positioned across the membrane to interact with components in the extracellular space and the periplasm," Buchanan says.
She says such structural details are the key to molecular understanding of transport and maybe even the design of some new vaccines.
 |
|
Reinhard
Grisshammer
|
G-Protein–Coupled Receptors
Reinhard Grisshammer, a staff scientist in NIDDK’s Laboratory of Molecular Biology, studies the enormous family of G-protein–coupled receptors (GPCRs). About 1,000 GPCRs have been identified, 300 to 400 of which are found throughout the body binding endogenous ligands (the remainder are chemosensory GPCRs for odors, pheromones, or tastes).
Recognizing signaling molecules like epinephrine and neuropeptides, GPCRs "are, of course, major drug targets for disease therapy," Grisshammer says. But his interests lie in understanding "the basic mechanisms of how these receptors see their ligand, how they change conformation, and how they transfer the signal across the membrane and lead to activation of a heterotrimeric G-protein."
A crystal structure for only one mammalian GPCR is known—bovine retinal rhodposin. Kryzysztof Palczewski’s group at the University of Washington took many years to solve its crystal structure.
For the receptors Grisshammer is interested in studying, which are much less abundant than bovine rhodopsin, "you need to do the tedious expression, purification, and then crystallization. And so we have worked for 15 years on developing methods to express membrane proteins, including GPCRs, in functional form and to establish robust purification schemes at large scale."
In collaboration with Joseph Shiloach at NIDDK’s Biotechnology Unit, Grisshammer uses a 200-liter fermentation tank to obtain receptor material of high quality for the regular purification of receptors.
These techniques have been honed to the point that he can produce 5 mg of functional receptor molecules each week. These lots are used to set up crystallization experiments and to develop specific antibodies that aid crystallization.
While scientists believe that the overall structures of GPCR families—for example, the rhodopsin family—–are similar, small differences in the ligand-binding regions may determine how tightly those ligands bind. "That’s the exciting bit, actually, so having structures from a few representatives would be quite good," Grisshammer observes.
P-Glycoprotein
 |
|
Di
Xia
|
Di Xia, an investigator in NCI’s Laboratory of Cell Biology, focuses his crystallographic efforts on proteins responsible for multidrug resistance. "Resistance is everywhere," Di Xia says.
Xia’s main focus is on P-glycoprotein (Pgp), the product of the MDR1 (multidrug resistance) gene.
First cloned by Ira Pastan and Michael Gottesman at NCI in the mid-1980s, Pgp belongs to a family of transporters that includes the gene for cystic fibrosis. Pgp is especially important in cancer management: Once selected in one anticancer drug, cells express the Pgp gene and pump many clinically important drugs, including anticancer agents and HIV protease inhibitors, out of the cell. This ATP-requiring pumping action reduces the intracellular concentration of drugs below their effective dose.
Because cancer cells appear much more sensitive to the function of Pgp than do normal cells, drugs designed to block Pgp could be extremely useful in the clinic, Xia remarks. (According to CA: A Cancer Journal for Clinicians, in 2001, there were more than 1.2 million new cancer cases in the United States and more than half a million deaths, most the consequence of chemoresistance.)
How Pgp captures and removes diverse drugs is not known. Xia’s lab is pursuing this by seeking to understand how the protein works at an atomic level.
In collaboration with Suresh Ambudkar at NCI, Xia began by expressing and purifying human Pgp. Several common expression systems lack the cellular machinery to produce, modify, and transport human Pgp to membranes and therefore do not generate protein.
Only in insect cells could human Pgp be generated in reasonable amounts, but the yield has been low, and larger cultures have proved less efficient for protein production than small ones. Additionally, purification protocols need to be tweaked. "This is risky business," Xia says.
There are many hypotheses as to why high-level expression of membrane proteins is difficult. One of them is membrane crowding.
To address the problem, Xia is using a novel system that places expression of transporter genes under the control of bacterial photosynthesis promoters to take advantage of the ability of photosynthetically grown bacteria to produce large amounts of fresh membranes.
While developing innovative techniques for expressing human Pgp, Xia is also working on bacterial and yeast Pgp homolog proteins. He hypothesizes that Pgps from all three groups will have similar structures and mechanisms of action.
Bacterial and yeast proteins are easier to study because cranking up expression of native Pgp is easier than modifying the yield of human transgenes. Xia’s group can now purify 200 mg of protein using NIDDK’s 200-liter fermentation tank—sufficient to begin growing crystals. With crystals in hand, Xia will test his hypothesis by determining the similarity of bacterial and yeast Pgps
 |
|
Mark
Mayer
|
Brain Receptors
Mark Mayer, head of NICHD’s Section on Neurophysiology and Biophysics, only recently tried his hand as a crystallographer. Trained as a neuroscientist, he has been studying the functional properties of glutamate receptors for more than 20 years, "way before they were cloned."
Found throughout the mammalian central nervous system, glutamate receptors transfer excitatory signals between cells by allowing sodium to enter a cell in response to the binding of a molecule.
Glutamate receptors "do all kinds of wonderful things in the brain," Mayer notes, indicating that his goal is to understand how they work at the molecular level. Mayer says ligand binding is "still quite difficult to model . . . until you’ve got really detailed side-chain information for a particular subunit."
Mayer currently works on kainate receptors, one of four members of the glutamate receptor family. Biochemical studies indicate that the protein has four domains, and it’s believed that four protein molecules—each with 1,000 amino acids—combine to form a functional kainate receptor.
This large size and degree of complexity make it impossible to study the intact, functional channel. "We’ve resorted to protein engineering to generate soluble constructs of interesting bits of the molecule," Mayer says.
The bit on which his lab is currently focused is the ligand-binding core. This domain determines subtype selectivity for substrates. "The receptors that have functional roles in the brain are strikingly diverse," Mayer says. "That’s due to small numbers of amino acid substitutions having very, very dramatic effects on selectivity."
The ligand-binding core can be thought of as a clamshell, with the ion channel separating the two halves of the shell. Linkers were designed to excise the membrane-spanning region and tie the two halves together. The product is a soluble protein that still binds ligand, implying that the structure has not been perturbed.
This approach works because "these multidomain proteins have quite clear domain boundaries," Mayer says. "One can excise the portion of the molecule that is selective for the ligands."
The engineered eukaryotic protein can be expressed in bacteria. "It’s slightly cumbersome juggling 12 liters" of cultures in shaker flasks, but, Mayer notes, "growing bacteria is really not a problem."
His lab has expressed, purified, and crystallized two members of a low-affinity kainate subfamily that have "striking differences in binding properties." In fact, he remarks, "the drug industry is very interested in these because they are potential analgesic and anticonvulsant targets.".
 |
|
Joseph
Mindell
|
Chloride Channels
Joseph Mindell, an investigator in the NINDS Membrane Transport Biophysics Unit, applies a range of methods to examine how chloride channels (ClCs) work. Broadly expressed in tissues, ClCs play roles as diverse as salt transport, brain function, muscle activity, and maintenance of intracellular pH.
For historical reasons, the ClC family wasn’t well studied, but that’s changing, Mindell says. He developed a structure of a ClC using electron microscopy and is now "going back to look at function in light of structure."
Mindell is taking a two-pronged approach to this goal. Little is known about which—and where—portions of bacterial ClC molecules move during chloride ion transport. Mindell’s lab is addressing this by attaching fluorophores to different regions of the protein. The fluoresence characteristics of the fluorophores change depending on the hydrophobicity of their environment.
Armed with this information, Mindell can then create, express, and eventually crystallize and solve X-ray structures for mutants "to get insight into how the whole thing works," he says. "The bacterial system is a great system for that," he notes. "We can make as much [ClC] as we want, and it’s easy to manipulate in the large quantities that we need for these sorts of classical biochemical experiments."
Mindell says that while he was focusing on structure, lots of fascinating "biology has emerged," including indications that proteins belonging to at least one of three ClC families are located on the intracellular membranes of lysosomes.
The development of new methods for measuring ion currents from currently inaccessible places, such as lysosomes, is the second prong of Mindell’s research program.
"The hope is to broaden the number of channels that we can actually look at and therefore start to look at the conserved properties of the family vs. things that might be specific to [just] one or two. And in the meantime, we may learn something about how the channels in lysosomes work."
New Approaches To Structure
Although the structures of even large membrane proteins can be solved by X-ray crystallographic techniques, these snapshots have key limitations beyond crystallization.
For example, movement is an inherent property of functioning transporters, receptors, and channels. A crystal, however, is a static structure; proteins must be locked into one, and only one, conformation for X-ray crystallographic analysis.
 |
|
James
Hurley
|
Structural Genomics
One critical class of molecules, amphitrophic membrane proteins, resides in membranes only when they’ve been activated. James Hurley, a senior investigator in NIDDK’s Laboratory of Molecular Biology, says "a great deal of signal transduction involves signaling enzymes that translocate from the cytosol to membranes upon activation." Hurley’s group studies these proteins, which are involved in trafficking as well as signalling.
Hurley’s undergraduate training was in particle physics. He developed his interest in signal transduction during his doctoral training. Upon arriving at NIH in 1992, he began his research program by "opening up Stryer [Lubert Stryer, Biochemistry] and looking at what was downstream of receptors."
Hurley’s lab went on to crystallize and solve structures of important domains from such classical second messengers as protein kinase C, phospholipase C, and adenyl cyclase.
Hurley’s lab has now moved from looking at protein domains to tackling protein complexes using a structural genomics approach. Thus, instead of looking at individual molecules, his lab will now be surveying and comparing domains and structures of whole protein families.
"A lot of trafficking is carried out not by single domains but by complexes that fold together," he observes. He is working in collaboration with Juan Bonifacino’s group in NICHD to study sorting into endosomal pathways. Hurley has also begun to look at other protein complexes involved in subcellular targeting and membrane trafficking.
Electron Microscopy
 |
|
Sriram
Subramaniam
|
Sriram Subramaniam, chief of the Section on Biophysics at NCI’s Laboratory of Biochemistry, is focused on developing and refining an alternative imaging modality: high-resolution electron microscopy.
The immediate product of a high-resolution electron microscopy experiment is an image of a protein or protein crystal that contains three-dimensional data compressed into two dimensions, much like an X-ray image in a computed tomography (CT) scan. Subramaniam’s lab uses a suite of techniques to extract the information present in these images to reconstruct the structures of proteins, protein complexes, organelles, and even whole cells.
Electron Crystallography. One of the techniques being used is electron crystallography. Membrane proteins can be crystallized in the plane of lipid bilayer to form two-dimensional crystals.
"The goal of electron crystallography," Subramaniam says, "is no different from that of X-ray crystallography. We want to solve atomic structures." However, because the protein is in a lipid bilayer, it is closer to its native environment than it is in the 3-D crystals used for X-ray crystallography.
This special feature provides an opportunity to study physiologically meaningful conformational changes using electron crystallography.
By analyzing and comparing diffraction patterns of 402 different crystals of the proton pump bacteriorhodopsin, each pattern taken at a slightly different angle, Subra-maniam was able to do just that and describe the structure of the protein caught in the act of pumping a proton across the membrane.
"We now have a detailed understanding of how a proton goes across the membrane, how it is pumped. What these structures show is that opening of the cytoplasmic side allows the helices to move and the proton to come in."
A similar approach is also being used in his lab to generate a high-resolution structure of the 12-helix membrane protein that transports oxalate (see graphic).
Electron Tomography. Another technique Subramaniam’s group is using to peer at the organization of membrane proteins in vivo is electron tomography (ET). ET is similar in concept to the CT used in clinical imaging, with one difference—the sample is moved relative to the electron beam in ET, whereas the beam is moved relative to the object in CT. ET is a powerful method for visualizing at molecular resolution the structures of things such as whole cells that cannot be crystallized in 2-D or in 3-D.
One of the projects Subramaniam’s lab is using ET to pursue is bacterial chemotaxis.
Bacteria possess an elaborate collection of protein machines that can direct the movement of the bacterium in response to molecules detected at the cell surface. The ultimate goal of using ET is to describe the structure and changes in organization of the molecular apparatus as it swings into action to generate the bacterium’s response.
By combining previously published X-ray data with tomographic analyses of fixed cells, Subramaniam’s team has observed and analyzed the arrangement of the serine receptor, Tsr, in intracellular membranes. "We discovered that these proteins form these remarkable assemblies inside cells, which we called zippers," Subramaniam recounts. The observation of this novel form has led the lab to propose a new picture of the mechanisms by which bacteria respond to external stimuli.
. . . and More. Another technique that Subramaniam and colleagues are pursuing in collaboration with Jacqueline Milne, also of NCI, involves averaging images from thousands of individual molecules to generate 3-D structures of large protein complexes that cannot be easily crystallized. The team recently reported the molecular structure of pyruvate dehydrogenase, a 10-megadalton protein complex, using these approaches.
Despite these advances, "it’s still very early days," Subramaniam says. He is setting ever more ambitious goals—and knows even more powerful techniques will be needed.
For example, his lab is trying to develop new methods of identifying specific proteins in a cell, much like green fluorescent protein is used today—only at "10 to 100 times greater spatial resolution. In the end, we want to be able to take a volume and [get] a three-dimensional localization of all molecules of a particular kind," he says.
To that purpose, the lab is developing new tools for automation of data collection and processing.
Rising to the Challenge
Technical advances notwithstanding, the continuing problems with expression, size, complexity, and purification add up to enormous investments in time and money to achieve structure determination of membrane proteins, NIH’s membrane protein masters say. Nevertheless, they are uniformly undaunted. All note that the generous and stable funding of the intramural program increases their productivity. Grisshammer says that one can "really go full speed" at NIH.
One key to their progress is that, although scattered among various institutes, the structural biologists share expertise to address technical difficulties. Buchanan notes that the wide variety of scientists at NIH is one of its strengths. Grisshammer concurs, saying that at most institutions "you have to solve the problems by e-mail or telephone. Here, you can just go and ask for help or advice or materials."
To spread NIH’s experience even further, Grisshammer has organized a membrane protein scientific interest group that meets regularly and has grown to more than 60 members.
As complex, wiggly, mysterious, rare, hard to pin down, and risky as they might be, membrane proteins remain well worth the challenge to the scientists the Catalyst interviewed. Grisshammer proceeds "without fear or hesitation." Mindell finds it "fun to work on problems where you don’t know the answer."
And Xia is sure that he
and his colleagues have the right approach: "We know what we are doing,
avoid problems, and solve problems one by one. Eventually it should work."
![]()