
| T H E N I H C A T A L Y S T | M A Y – J U N E 2008 |
|
|
|
Heart Study Moves to the IRPFRAMINGHAM, MARYLAND? |
by Christopher Wanjek |
 |
Framingham Heart Study Director Daniel Levy
|
In 1948, as death rates from cardiovascular disease
continued their gradual yet
steadfast rise from decades prior and were more than a little worrisome, the
National Heart Institute established a novel study in the historic town of
Framingham, Mass.
Little was known then about the
general causes of heart disease and stroke. The Framingham Heart Study, a
longitudinal study originally composed of 5,209 Framingham adult residents,
ultimately established most of the risk factors that are now embedded in the
vernacular: high blood pressure,
high blood cholesterol, smoking, obesity, diabetes, and physical inactivity.
 |
Associate Director Christopher O'Donnell |
To embark on a
long-term study of thousands of people who had no overt symptoms of
cardiovascular disease was an ambitious project not without its share of
criticism. Early peer-reviewed papers from the Framingham research team mostly
justified the epidemiological proach to studying heart disease.
But by the late 1950s, the study was
bearing fruit, as researchers began to see the effects of high blood pressure
and cigarette smoking on the hearts of their subjects, who returned every two
years for a detailed examination.
Dozens of landmark papers followed.
Framingham, the city, soon became synonymous with the study. In 1971, the study
enrolled a second-generation cohort—5,124 of the original participants’
adult children and their spouses. Then the grandchildren joined in 2002. Throughout this period the study
enjoyed the continuous support of NIH extramural funding.
Beating Stronger Every Day
Sixty years and nearly 2,000 journal
articles later, the NHLBI’s Framingham research team has joined the NIH
intramural program. The study is still within NHLBI, only now it has reinvented
itself once again. Indeed, the Framingham Heart Study remains as innovative and
promising as it was a half-century ago.
The impetus for the move was to
“leverage 60 years of data collected at Framingham” with intramural resources,
such as in gene-expression profiling and bioinformatics,” in order “to make the
investment all the more cost effective,” said Daniel Levy, who joined the study
in 1984 and became its director in 1995.
Levy has numerous projects under
consideration that can best be done in-house. Entering a feasibility stage this
June is a study to correlate gene expression with phenotypes and 500,000
genotyped SNPs in Framingham participants, using microarrays (perhaps better
described as “macroarrays”)—a 96-sample peg plate instead of the standard
chip. NHLBI has the core facility to undertake this project in the Clinical
Center.
 |
|
Modern probes of the heart: The NHLBI Genomics Core can study the gene expression profile of 96 samples at once on one plate. Facility director Nalina Raghavachari holds one such plate |
“We’re looking for new biology,”
said Peter Munson, head of the Mathematical and Statistical Computing
Laboratory within CIT’s Division of Computational Bioscience, who shifted his
attention to the Framingham project a year ago to develop data-analysis
software. Munson describes Framingham’s new focus as “a melding of good,
old-fashioned science and people with lots of technical know-how.”
Genome of the Heart
Framingham hinted at its new direction
in recent years with several large-scale genotyping projects, such as a
genome-wide scan of nearly 100,000 SNPs from 1,345 study subjects, using the
Affymetrix 100K Genechip.
Digging deeper, NHLBI launched the
Framingham SNP Health Association Resource (SHARe) project in early 2007, under the leadership of Christopher O’Donnell, Framingham’s
associate director and SHARe’s scientific director.
They upped the ante, too, with the
goal of genotyping approximately 550,000 SNPs in more than 9,000 participants
from three generations,
encompassing more than 900 families.
Stored within NCBI’s dbGaP and, as
its acronym signals, open to scientists worldwide (beginning in October 2008),
the SHARe database will contain all previous Framingham SNP and microsatellite
genotyping, as well as extensive phenotype information from the three-generation
cohort: quantitative measures such
as systolic blood pressure, total and HDL cholesterol, fasting glucose, and
cigarette use; anthropomorphic measures such as body mass index; biomarkers
such as fibrinogen and C-reactive proteins; and electrocardiography measures
such as the QT interval.
SHARe contains the 500K-SNP data and
may possibly house much more.
 |
Core scientists: (left to right) facility director Nalina
Raghavachari, technologist Kimberly Woodhouse, and research biologist Poching Liu |
Using the SHARe resource, O’Donnell
hopes to discover new genes underlying coronary heart disease and subclinical
atherosclerosis detected by computed tomography and other imaging measures.
Levy hopes to discover genes involved in hypertension and altered vascular
function. The project might relate
common genetic variation to alterations in gene expression as a means to get
one step closer to understanding functional changes in human DNA.
“The focus is on discovering new
genomic and genetic risk factors to identify the specific genetic sequences
underlying associations seen previously and to test how these new genetic
risk factors might be used to predict and prevent cardiovascular disease,” said
O’Donnell, who joined the Framingham Heart Study in 1996 and, like Levy, joined
the intramural program in 2007 as a tenured investigator in NHLBI who maintains
his base in Massachusetts.
“[Framingham] is a study that
reflects the real world,” he said, and SHARe brings that study to the world by
allowing scientists to compare genes within the Framingham study and between
similar heart studies.
New Expressions
The next level, as Levy sees it, is
discovering biomarkers and related therapeutics via a “phenomic” analysis of
gene expression—to link proteins and metabolites to risk factors.
Enter the Genomics Core, a facility
on the 8th floor of the CC to study gene expression, initiated by Eric Billings,
head of Bioinformatics and Systems Biology in NHLBI’s Intramural Research
Program, and now under the direction of Nalini Raghavachari.
This facility has automated the
sample-preparation protocol, allowing for a tremendous increase in capacity
while reducing noise to a 15 percent coefficient of variation. A robot can manipulate an “array of
arrays,” processing 96 samples at once, reducing batch effects, and exerting
exogenous controls.
Beginning this summer for about two
months, the Genomics Core will become an assembly line to study Framingham
samples. The feasibility aspect is to assess which types of biological samples
would be most useful in this mRNA analysis. Handling the sheer number of
samples once the project moves forward—estimated to be at least 7,000
samples—is less of a concern, although this is the largest project by far
that the Genomics Core has undertaken.
The Genomics Core processes about
1,000 samples a year; its largest single project has been a heart study for
NHLBI Director Elizabeth Nabel involving 200 samples. “This blows away anything
we’ve done before,” said Mark Gladwin, former chief of NHLBI’s Vascular
Medicine Branch, who coordinated this and other projects between NHLBI and the
CC.
Levy speaks eagerly of the enormous
research potential for Framingham within the intramural program, anticipating
biomarker discoveries that are proteomic, metabolomic, and lipomic. He’s
enthused about combining genetic and genomic information and applying systems
biology approaches on a population level, as well as about the possibility of
internal collaborations and recruiting high-quality fellows.
Genome-wide association studies have
proven themselves successful for diabetes and several inflammatory diseases,“
but the Framingham study can go far beyond that,” said O’Donnell.
![]()
 |
 |
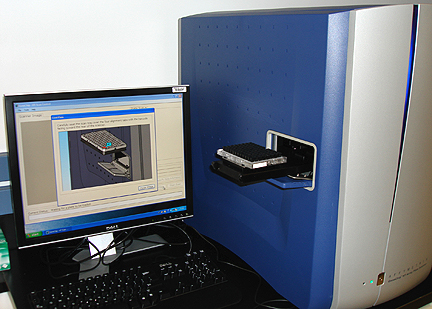 |
The Gene Chip Array Station robot can process and analyze
96 RNA samples at once within a few hours
|
Affymetrix fluidic stations, which can wash and stain
gene chips in an automated manner, are used in the Genomics Core to maximize
efficiency
|
The 96-sample plate that has been hybridized to 96
different RNA samples, washed and stained with fluorescent dye is scanned by
the laser scanner to detect expressed genes in the samples
|