
| T H E N I H C A T A L Y S T | S E P T E M B E R – O C T O B E R 2007 |
|
|
|
SIGNAL DISCOVERY OPENS PATHWAY TO THERAPIES FOR IRON-OVERLOAD ILLS |
by Christopher Wanjek |
 |
|
Iron-clad
team:
(left to right) lab chief Jeff Miller, senior author; postdoc Toshihiko
Tanno, lead author; and lab manager Terry
Lee, co-author, seated in front of computer display that summarizes
more than one million data points describing gene activity during human
erythropoiesis
|
Too much iron will leave a body frail, destroying joints, scarring the liver, fueling cancers and heart disease, and sapping vitality. These effects contradict everything we have learned from the nautical sage Popeye.
The human body has no means to shed excess iron aside from blood loss. So it regulates iron’s uptake from food through the peptide hepcidin, which hinders iron absorption via enterocyte cells in the gut.
The basics are well known. In the past year, however, Jeff Miller’s group in NIDDK’s Molecular Medicine Branch has reached a breakthrough in understanding the underlying chemical signaling that controls iron regulation, which could lead to diagnostic tools and possible treatment for those with iron-overload diseases such as thalassemia.
Miller, chief of NIDDK’s Molecular Genomics and Therapeutics Section, describes this research in the September 2007 issue of Nature Medicine, with lead author Toshihiko Tanno from his lab and other scientists from NIH and elsewhere.(1)
The team speculates that the newly discovered signal, involving growth-differentiation factor 15 (GDF15), might be common in many states of anemia and blood disorders as well as in liver disorders and some cancers.
"While what we were studying was a very unusual mechanism in the erythroid cells, what we may have discovered is a very generalizable mechanism of iron regulation, or of how tissue damage can activate certain hormones, whether it be from cancer or from liver disease or, in our case, from anemia," Miller said. "One discovery in a patient with thalassemia is now leading to other discoveries in other patients."
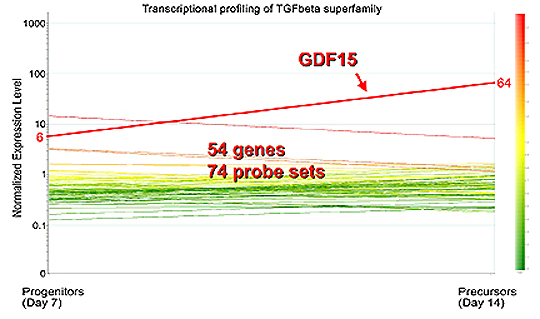 |
Identification
of GDF15 as the candidate molecule:
After transcriptional profiling of the TGF-b
superfamily members in erythroblasts, GDF15 stood out among 54 genes.
GDF15 suppresses the iron regulator hepcidin, and later it was revealed
that patients with thalassemia demonstrated 10-100–fold increases
in serum GDF15 levels when compared with healthy volunteers |
The discovery capitalized on the unique blend of resources at NIH, such as expertise in bioinformatics, the NIH Intramural Sequencing Center (NISC), collaborations with smart lab neighbors, and patients at the Clinical Center.
From Dinner to Bench to Bedside
Miller, a hematologist, studies a variety of blood diseases. The core of his lab’s research since the mid-1990s has been to map gene expression in the erythroid lineage.
"We have spent a decade trying to put together an infrastructure for all of the genes that are being turned on and off as an adult stem cell becomes a red [blood] cell," Miller said. "This information database allowed us to correlate clinical observations with animal models and allowed us to ask what signals could possibly regulate iron."
What hooked Miller on iron was a conversation about a year ago at a dinner with the local chapter of the Cooley’s Anemia Foundation, an advocacy group with members suffering from this form of thalassemia.
Cooley’s anemia is an autosomal-recessive blood disease affecting only about a thousand people nationally, although single-gene carriers for thalassemia are found in 10 percent or more of the population in Thailand, Cyprus, and other malarial regions. People with thalassemia are unable to properly synthesize one of the globin chains that make up hemoglobin and are thus prone to anemia. They have elevated numbers of erythroblasts in their bone marrow but reduced numbers of healthy erythrocytes circulating in the blood. Cooley’s anemia and other forms of thalassemia are treated primarily by frequent blood transfusions.
The advocacy group told Miller its greatest concern was iron overload, a main underlying cause of death for those with thalassemia. The excess iron can stunt growth, destroy the thyroid gland, damage the liver, and induce diabetes, to name a few complications. Doctors have known that, even in the absence of transfusions, thalassemia patients often accumulate iron—and transfusions markedly worsen this.
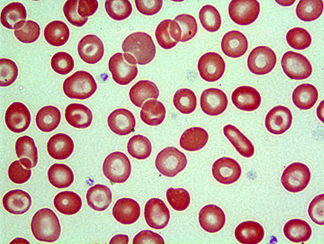 |
Bull’s
eye: Peripheral blood smear from a donor with thalassemia trait captures
classic telltale "target cells"—red cells that have a
central bull’s eye |
"Maybe there is something about the way they make their blood—incorrectly—that makes them absorb too much iron," Miller hypothesized.
Miller had hints from several sources—including the clinic he established at the Clinical Center and an animal model from the neighboring lab of Chuxia Deng, chief of the NIDDK Mammalian Genetics Section—that the signal could be a member of the transforming growth factor–b (TGF-b) superfamily of genes. He was particularly inspired by Deng’s work, which he had heard at a NIDDK retreat, and began to concentrate on TGF-b proteins, including GDF and bone morphogenetic proteins, all in the superfamily. He was also inspired to pursue the project by Carolyn Philpott, senior investigator in NIDDK’s Liver Diseases Branch and an expert on iron biology.
Alan Schechter, the NIDDK Molecular Medicine Branch chief, described Miller’s lab’s process of narrowing in on the suspected genes as going from "bedside to bench to bedside," one of the true advantages of the intramural research program.
"Jeff established two years ago a clinic in the hospital to see patients," Schechter said. "Once you have the robust clinical base, then you can go back and forth. . . . It’s an iterative process. You can’t just think some profound thought in a laboratory and then come over to the hospital in the middle of the night to apply it to patients."
Erythroid Gene Clearinghouse
 |
Susan
Leitman |
 |
Gerard
Bouffard |
 |
Alan
Schechter |
Tanno, an IRTA postdoc fellow, began the project with the meticulous process of identifying all the signals made by adult stem cells during their transformation from erythroblasts to red blood cells. Key to this work was the warehouse of data from the Human Genome Project and bioinformatics tools to analyze these data. A variety of bioinformatic approaches was used, including those available through NCBI, commercial software for array analyses, and custom programming strategies crafted by Gerard Bouffard of NHGRI, director of the NISC Bioinformatics Group.
The process entailed isolating mRNA from erythroid precursor cells, reverse-transcribing this to cDNA, creating thousands of expressed sequence tags (ESTs), removing contaminants, and then "BLASTing" them using NCBI’s BLAST tools to compare them with other gene sequences.
"BLAST is one of those tools you can’t imagine modern life without," said Bouffard, who wrote a complementary software program for Miller’s group "to distill out gold nuggets" from the BLAST results. He has worked with Miller on this project for several years.
All this information is posted at NIDDK’s Hembase, a public repository of information on erythroid ESTs and also sequences encoding several hundred additional genes with known expression in erythroid cells, organized and linked according to the location of these sequences within the human genome.
From Hint to Confirmation
Guiding Tanno on his needle-in-a-haystack search for an iron regulator in the midst of all these sequences was the clinic- and lab-driven hypothesis that iron regulation involved the TGF-b superfamily. He examined adult human erythroid cells from clinic patients to see whether any TGF-b genes were expressed.
Three candidates emerged from a long list, with GDF15 in particular copiously expressed by erythroid cells.
Going back to the clinic, the team checked whether patients with thal-assemia had too much GDF15 in their blood. The answer was as clear as a bell.
"They had incredibly high levels," Miller said, "a logarithmic increase in the level of this in their blood" relative to the patient’s iron overload and compared with healthy patients. "We were thinking, wow. . . . It was very satisfying to be able to go back to the clinic to these patients who were donating their blood to figure out if our hints in the laboratory were useful."
Once the group found what was increased in the blood, they could determine the mechanism: GDF15 appears to be an inhibitor of hepcidin, a major iron regulator made in the liver. This hepcidin suppression leads to more iron absorption.
"Since erythroblasts need iron to make hemoglobin, we reasoned that the increased number of erythroblasts in thalassemia may send stronger messages to the liver to suppress hepcidin and thereby absorb more iron even in the condition of iron overload," said Tanno.
Broader Implications
Miller has about a dozen thalassemia patients in his clinics, but he has been able to strengthen his ongoing research on this topic by studying blood from other patients, including those with other iron-overload diseases seen by Susan Leitman, chief of the Blood Services Section of the CC’s Department of Transfusion Medicine, as well as by receiving samples from thalassemia patients in Thailand, where nearly 1 percent of the population has some thalassemia disease.
"Many experts in iron regulation have postulated the existence of an ‘erythroid regulator’ of iron absorption for a while now,"said Leitman. "The search for this erythroid regulator has been a hot issue in labs focused on iron homeostasis. Jeff’s identification of GDF15 is a major breakthrough in this field."
The Thai samples came courtesy of Suthat Fucharoen of Mahidol University in Nakornpathom, Thailand, who had spent a summer at NIH several years ago and who has continued to collaborate with Miller, Schechter, and NIDDK Director Griffin Rodgers.
"This is the basis of how science works—exchanging visits and established collaborations," Schechter said, adding that Fucharoen’s collaboration has greatly aided in the discovery of iron regulation.
There are other anemias that don’t behave like thalassemia but still might be regulated by hepcidin and GDF15, Miller said. He also speculates that the involvement of GDF15 in thalassemia and other diseases, including cancers, may extend beyond the regulation of hepcidin.
"We’re hoping now—very much so—we can use this for diagnostic, prognostic, and even therapeutic purposes, because the discovery of signals immediately leads to the possibility of blocking those signals," Miller said.
"Information that has come from the study of hemoglobin diseases has been applicable to lots of other diseases," said Schechter, citing Linus Pauling’s 1949 paper defining sickle cell as the first known molecular disease. What Miller has done, Schechter added, was make "use of the new era of genomic medicine to define a subfield that probably could be called human erythroid cell genomics . . . to pioneer a whole new field within hematology."
Whereas the current project has been focused almost entirely on thalassemia, the lab hopes to apply this translational approach to other diseases and to ensure that the broader scientific community has access to their genomic studies to facilitate basic and clinical connections that would otherwise be difficult to make.
"All this information
can now be organized because of the Human Genome Project. Let’s not forget
that," Miller said. "Now we’re taking the next step—and
perhaps the more difficult though most important step—to use the information
to make people feel better." ![]()
1. T. Tanno, N.V. Bhanu, P.A. Oneal, S.-H. Goh, P. Staker, Y.T. Lee, et al., "High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin," Nature Medicine 13, 1096–1101 (2007); published online, August 26, 2007.