| T H E N I H C A T A L Y S T | J U L Y – A U G U S T 2007 |
| | |
Bethesda Makes Another Name for ItselfCC, NIAID RESEARCHERS UNCOVER NEW DISEASE-CAUSING BACTERIUM |
|
Bethesda, the famed healing bath of ancient Jerusalem and present-day home in Maryland to a certain collection of health institutes, has acquired one more historic attribute: There's now a bacterium named after it.
The newly identified bacterium is called Granulibacter bethesdensis, a tribute to the neighborhood where the discovery was made.
After a three-year investigation, researchers at NIAID and the CC determined that this bacterium was the cause of lymphadenitis in a patient with the rare genetic immune disorder known as chronic granulomatous disease (CGD).
The finding underscores bacteria-disease connections that still await discovery.
It took some sleuthing. G. bethesdensis is difficult to culture and is not only a new species but constitutes a whole new genus in a family of acetic-acid-producing bacteria never before associated with human disease. The bacterium's close cousins are used commercially in the production of vinegar.
The discovery also highlights the unique combination of talent and resources at NIH, in particular the close relationship among the CC's microbiologists and clinical investigators, who typically visit the microbiology lab every day to discuss their patient's infections.
"People are finding new bacteria all the time," said David Greenberg, an associate clinical investigator in the NIAID Laboratory of Clinical Infectious Diseases (LCID) and lead author on two papers describing the new bacterium.

Scanning electron micrograph of a neutrophil about to engulf a Granulibacter bethesdensis cluster. Image courtesy of Dave Dorward, NIAID
A Rare Find
"But to find an organism that actually causes human disease, albeit in an immunocompromised population, is rare. That's what makes this very exciting."
Greenberg and his colleagues, led by Steve Holland, LCID chief, are now trying to understand what makes this bacterium virulent to CGD patients and whether it is implicated in other, more common diseases.
They also hope to determine what the immunodominant antigens are in order to develop a screening test. Such CGD research has provided insights into the nature of inflammation and immunology.
People with CGD are susceptible to serious infections of the lungs, lymph nodes, skin, and bone from seemingly unrelated bacteria and fungi. The CC is the leading research facility studying CGD, which affects about one in 250,000 people worldwide.
The CGD patient, who has since largely recovered but continues to be monitored, was first referred to the CC in 2003 after his doctors in his hometown could not determine the cause of his swollen lymph nodes.
 |
|  |
| 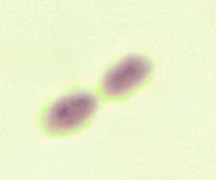
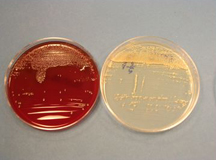
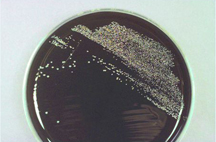
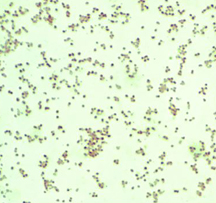
The New Kid in Town: (top) A buffered-charcoal yeast-extract plate showing the very first isolate of Granulibacter bethesdensis; a gram stain revealing a gram-negative coccobacillus, but standard microbiology laboratory procedures cannot identify the bacterium; a pair of the newly discovered bacterium up close; (bottom) gene sequencing and biomedical profiling reveal bacterium family, and further chemical tests [on right] show the bacterium can grow on methanol, a clue to its natural environment. Images courtesy of Adrian Zelazny, NIH.
Step 1: Culturing
Biopsies of the patient's lymph nodes were sent to the microbiology lab, where Alexandra Wong, a medical technologist in the CC's Department of Laboratory Medicine, first discovered the bacterium growing in cultures of the biopsies.
Because many patients who come to the CC are immunocompromised and susceptible to a wide variety of infections, culture conditions are optimized for detection of bacteria that grow slowly or are fastidious. This was fortunate because this bacterium took five days to grow, about twice as long as most bacteria.
Traditional laboratory staining demonstrated this was a gram-negative bacterium, but standard biochemical tests couldn't pinpoint its identity. For many labs, had they even been able to culture this bacterium, this point would have been the end of the road.
The CC-NIAID team, however, was confident that it was dealing with something unique and not simply a lab contaminant. The same bacterium was isolated from multiple biopsies, and gram-negative bacteria are not common lab contaminants.
So after consulting with Holland and Greenberg, Patrick Murray, chief of clinical microbiology at the CC, asked Adrian Zelazny, then a Fogarty fellow in the microbiology lab, to pursue other techniques for identifying the bacterium.
Step 2: Sequencing
Zelazny's first approach was to sequence the bacterium's 16S ribosomal gene, now a well-established test in the microbiology lab. These results were the first clue of the organism's uniqueness. Upon sequencing, the team found that the closest match was the Acetobacteraceae family, bacteria known to convert alcohol into vinegar.
"It still wasn't a very good match, so early on we had a sense we were dealing with something new," Greenberg said. "The question is, how do you figure out if you're dealing with a brand new species—or genus, for that matter?"
Zelazny, today a staff scientist in Holland's lab, subjected the mystery bacterium to more biochemical tests and compared the results to those from tests on the Acetobacteraceae family. The team found that the bacterium expressed unique biological signatures. He then performed a polyphasic taxonomic study, which looks at the sequence of many genes, not just 16S. The final tests included DNA-DNA hybridization, a technique to measure the genetic distance between the new bacterium and members of the Acetobacteraceae family.
"The bacterium emerged as a distinct branch," Zelazny said, and the combination of sequencing and biochemical tests "proved it was a new member in the Acetobacteraceae family."
The outstanding question that remained was whether G. bethesdensis was the culprit causing lymphadenitis. Once again, the CC could accommodate.
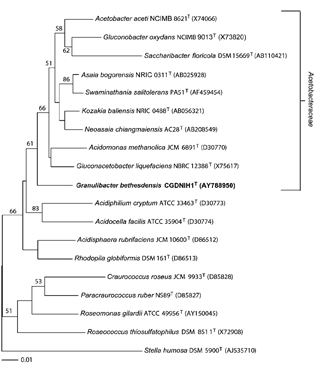
Sequencing of the 16s ribosomal gene revealed that Granulibacter bethesdensis constituted its own branch. Image courtesy of Greenberg et. al.
Step 3: Demonstrating Pathogenicity
Years ago, Holland's lab had created a mouse model for CGD, a well-studied inherited disorder of phagocytic cells caused by a defect in the phagocyte NADPH oxidase, an enzyme responsible for creating the reactive oxidant superoxide.
Li Ding, a senior research assistant in Holland's lab, exposed the enzyme-deficient mice to G. bethesdensis and, sure enough, the mice developed a pathology similar to that of the original patient. More important, she could re-isolate the bacterium and grow it again, demonstrating how G. bethesdensis multiplied in the mice.
Further proof came after the team placed the sequence information into GenBank and word spread about the bacterium. G. bethesdensis was isolated from two more CGD patients with lymphadenitis at the CC and from a fourth patient with lymphadenitis in Houston. Working with Frida Stock, a research assistant in the microbiology department, the group has found that all the organisms are genetically distinct.
Greenberg is summarizing the four case studies in a third paper. So far, G. bethesdensis has not been isolated from the environment, but similar organisms grow on sugar cane and tropical fruit, essentially high-sugar environments where natural fermentation yields alcohol for the bacteria to feed on. Three of the four patients are from sunny climes; one is from Panama. The original patient is from the north but developed his disease shortly after a visit to the Bahamas.
Many More Out There
"Why was [G. bethesdensis] discovered here?" Greenberg asked. "Probably because we have a spectacular microbiology lab. We have the means not only to identify things when they arise, but to take it to the next level and start asking questions: Is this really a pathogen, and how do you prove that?"
Murray attributed it to being "blessed with bright fellows" and "given the resources to pursue interesting research."
"There are a tremendous amount of diseases in the world for which there is no known etiology," Greenberg said. "And people have been suspicious that there may be an infectious connection. . . . At the end of the day there are disease-causing organisms that remain to be discovered. People should be on the lookout for them."

Lab Sleuths: (left to right, back row) Patrick Murray, Adrian Zelazny, David E. Greenberg, and Steven Holland; (front row) Li Ding, Frida Stock, and Alexandra Wong. Image courtesy of Ernie Branson, NIH
What's in a Name?
The naming of the new organism—carried out by Greenberg, Zelazny, Holland, and Murray—was nearly as tricky as its discovery.
The four liked the way Granulobacter bethesdensis rolled off the tongue, and, indeed, in the first published paper, issued April 2006, the organism went by that name. But by the time the second paper was issued, in November that year, they had been informed that Granulibacter, with an i, is the proper word to accompany the Latin masculine adjective bethesdensis—and the name was changed accordingly. Such strict rules make the naming process another difficult task in the discovery process.
Fungi are even harder to name, Murray joked.