| T H E N I H C A T A L Y S T | J U L Y – A U G U S T 2001 |
|
|
|
IN THE MAINSTREAM:MICROARRAY RIVULETS FOUND CAMPUS-WIDE |
|
 |
Microarray technology, a technique used for the rapid screening of genes using chips containing immobilized cDNA fragments, has burst upon the NIH intramural scene.
An Array of Users
The Microarray User Group, an NIH special interest group that began in 1998, grows weekly and is now approaching 300 members, almost half of whom are intramural investigators (others hail from academia and industry in this country and around the world).
The ranks are filled with investigators from nearly every institute, with the NCI cohort currently the largest.
According to Katherine Peterson of NEI, who heads the group, the range of experience among members encompasses "Affymetrix experts, who use oligonucleotides on a silicon wafer [and] those scientists who use spotted arrays on a nylon membrane." Members use the group to exchange ideas, listen to guest speakers, and share equipment.
Another element of the research involves the Center for Information Technology here on campus. Peter Munson, acting chief of the Mathematical and Statistical Computing Lab, notes that "there has been a realncrease in the sophistication and expectations of the users of this technology. We develop software for doing image analysis and preliminary statistics for P53-labeled arrays and for fluorescence-labeled glass microarray." The technology, he says, is "quite complicated, has limitations, and will require real work to apply to [complex] problems."
 |
|
Microarrayers:
Ed Liu (left), former NCI clinical director, and Lisa Gangi
|
 |
|
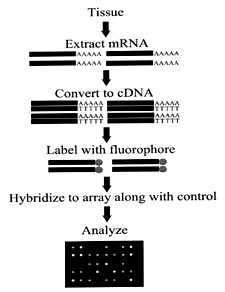
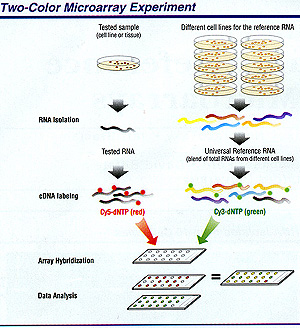
Microarrays
(or DNA chips) provide a rapid and simultaneous screening of many thousands
of genes, displaying which genes are expressed at that given stage in
the development of a cell. mRNA is isolated from cells at different stages
of development, converted to cDNA (using reverse transcriptase), and labeled
with fluorescent deoxynucleotides. The fluorescent cDNA can then be used
as probes, each hybridizing to complementary sequences on the microarray
chips. These chips are essentially glass slides coated with poly-L lysine
and contain immobilized PCR-amplified cDNA fragments. The time and details
of gene expression provide insight into a cell’s role at a particular
phase of cellular development. The patterns of gene expression are characterized
for their relevance to disease. Additionally, software developed by NIH’s
CIT aids in detection of subtle differences of gene expression.
|
Earlier this year, before then-NCI clinical director Ed Liu left NIH to head the National Genomic Institute in Singapore, he presented an expression-array sampler of recent laboratory findings in the IRP and their relevance to human diseases. Among them were the identification of molecular pathways involved in breast cancer development and the means to distinguish low- and high-grade ovarian cancers.
One Project
For Lisa Gangi—an NCI investigator with roots in Liu’s lab who recently moved out to the microarray research facility at NCI’s Laboratory of Molecular Technology in Frederick—microarrays are a "tool to look at thousands of genes at once, a snapshot of all gene expression at a given time."
Gangi has been using that tool to track normal gene expression in mouse breast development and the upregulation of proteins that accompanies pregnancy, lactation, and involution. Her findings in the mouse now inform her examinations of patients’ breast tumors. Specifically, she and her group are searching for correlations in the gene expression patterns of the DNA repair pathway seen in the mouse.
Gangi typically views 5,000 to 7,000 genes on any given glass slide and uses two-color probes to distinguish tumor from normal tissue in both mouse and human mammary samples. She has documented a tenfold increase in the expression of DNA repair pathway genes during involution compared with during lactation; she is now focusing on the identification of novel genes that are upregulated in the apoptosis pathway during involution.
She and her team have observed a decrease in survival factors—also known as anti-apoptosis genes, such as bcl-2, bcl-3, and ICH/IS—and a concomitant increase in pro-apoptotic genes, such as bcl-x, bax, and the matrix metalloprotease genes during the first 24 hours of involution. At late time points, there is a marked increase in the expression of adhesion molecule genes and other cytoskeletal genes required for cellular remodeling.
This upregulation of repair genes suggests that normal mammary KU 70/80 and ATM, which are involved in the repair of DNA double-strand breaks, are involved in both mammary gland nvolution and breast cancer development.
"It’s interesting,"
Gangi says, "that many human breast tumors appear to have aberrant levels
of DNA repair genes. . . . We hope to elucidate whether the increase in the
levels of these proteins is a normal consequence of cellular apoptosis and remodeling
and whether there is a connection in DNA repair to downstream events leading
to tumorigenesis." ![]()